Wine Quality Regression
Performing regression and binary classification on Wine Quality Databases in the UCI repository. This is a supervised learning project since there is a training variable.
We will utilize regularization on our linear regression to prevent overfitting, and we will utilize ensembles to improve our logistic regression and decision trees. Cross validation will allow us to tune our hyperparameters. I was able to achieve the classification win condition, but not the regression win condition
Regression win condition: the wine quality comes in integer numbers. We will attempt to predict quality to a >90% accuracy after rounding our predictions.
Classification win condition: The qualities range from 3-9, the mean is about 5.7-5.9 for both datasets, and the std is about .8-.9 for both. Any value 7 or above would be more than one STD above the mean for either dataset, we can define these as good wines and the others as bad and turn this into a binary classification problem. We will look for an AUROC score above .9.
0. Import Libraries
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
%matplotlib inline
import seaborn as sns
sns.set_style('darkgrid')
from sklearn.model_selection import train_test_split
from sklearn.linear_model import Lasso, Ridge, ElasticNet
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor, RandomForestClassifier, GradientBoostingClassifier
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import GridSearchCV
import warnings
from sklearn.exceptions import DataConversionWarning
warnings.filterwarnings(action='ignore', category=DataConversionWarning)
from sklearn.exceptions import NotFittedError
from sklearn.metrics import r2_score
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import accuracy_score
from sklearn.metrics import roc_auc_score
I. Exploratory Analysis
ex_red = pd.read_csv('winequality-red.csv', sep=';')
ex_red.head()
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7.4 | 0.70 | 0.00 | 1.9 | 0.076 | 11.0 | 34.0 | 0.9978 | 3.51 | 0.56 | 9.4 | 5 |
| 1 | 7.8 | 0.88 | 0.00 | 2.6 | 0.098 | 25.0 | 67.0 | 0.9968 | 3.20 | 0.68 | 9.8 | 5 |
| 2 | 7.8 | 0.76 | 0.04 | 2.3 | 0.092 | 15.0 | 54.0 | 0.9970 | 3.26 | 0.65 | 9.8 | 5 |
| 3 | 11.2 | 0.28 | 0.56 | 1.9 | 0.075 | 17.0 | 60.0 | 0.9980 | 3.16 | 0.58 | 9.8 | 6 |
| 4 | 7.4 | 0.70 | 0.00 | 1.9 | 0.076 | 11.0 | 34.0 | 0.9978 | 3.51 | 0.56 | 9.4 | 5 |
ex_white = pd.read_csv('winequality-white.csv', sep=';')
ex_white.head()
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7.0 | 0.27 | 0.36 | 20.7 | 0.045 | 45.0 | 170.0 | 1.0010 | 3.00 | 0.45 | 8.8 | 6 |
| 1 | 6.3 | 0.30 | 0.34 | 1.6 | 0.049 | 14.0 | 132.0 | 0.9940 | 3.30 | 0.49 | 9.5 | 6 |
| 2 | 8.1 | 0.28 | 0.40 | 6.9 | 0.050 | 30.0 | 97.0 | 0.9951 | 3.26 | 0.44 | 10.1 | 6 |
| 3 | 7.2 | 0.23 | 0.32 | 8.5 | 0.058 | 47.0 | 186.0 | 0.9956 | 3.19 | 0.40 | 9.9 | 6 |
| 4 | 7.2 | 0.23 | 0.32 | 8.5 | 0.058 | 47.0 | 186.0 | 0.9956 | 3.19 | 0.40 | 9.9 | 6 |
ex_red.isnull().sum()
fixed acidity 0
volatile acidity 0
citric acid 0
residual sugar 0
chlorides 0
free sulfur dioxide 0
total sulfur dioxide 0
density 0
pH 0
sulphates 0
alcohol 0
quality 0
dtype: int64
ex_white.isnull().sum()
fixed acidity 0
volatile acidity 0
citric acid 0
residual sugar 0
chlorides 0
free sulfur dioxide 0
total sulfur dioxide 0
density 0
pH 0
sulphates 0
alcohol 0
quality 0
dtype: int64
print(ex_red.shape)
ex_red.drop_duplicates()
print(ex_red.shape)
(1599, 12)
(1599, 12)
print(ex_white.shape)
ex_red.drop_duplicates()
print(ex_white.shape)
(4898, 12)
(4898, 12)
ex_red.dtypes
fixed acidity float64
volatile acidity float64
citric acid float64
residual sugar float64
chlorides float64
free sulfur dioxide float64
total sulfur dioxide float64
density float64
pH float64
sulphates float64
alcohol float64
quality int64
dtype: object
ex_red.describe()
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 |
| mean | 8.319637 | 0.527821 | 0.270976 | 2.538806 | 0.087467 | 15.874922 | 46.467792 | 0.996747 | 3.311113 | 0.658149 | 10.422983 | 5.636023 |
| std | 1.741096 | 0.179060 | 0.194801 | 1.409928 | 0.047065 | 10.460157 | 32.895324 | 0.001887 | 0.154386 | 0.169507 | 1.065668 | 0.807569 |
| min | 4.600000 | 0.120000 | 0.000000 | 0.900000 | 0.012000 | 1.000000 | 6.000000 | 0.990070 | 2.740000 | 0.330000 | 8.400000 | 3.000000 |
| 25% | 7.100000 | 0.390000 | 0.090000 | 1.900000 | 0.070000 | 7.000000 | 22.000000 | 0.995600 | 3.210000 | 0.550000 | 9.500000 | 5.000000 |
| 50% | 7.900000 | 0.520000 | 0.260000 | 2.200000 | 0.079000 | 14.000000 | 38.000000 | 0.996750 | 3.310000 | 0.620000 | 10.200000 | 6.000000 |
| 75% | 9.200000 | 0.640000 | 0.420000 | 2.600000 | 0.090000 | 21.000000 | 62.000000 | 0.997835 | 3.400000 | 0.730000 | 11.100000 | 6.000000 |
| max | 15.900000 | 1.580000 | 1.000000 | 15.500000 | 0.611000 | 72.000000 | 289.000000 | 1.003690 | 4.010000 | 2.000000 | 14.900000 | 8.000000 |
ex_white.describe()
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 |
| mean | 6.854788 | 0.278241 | 0.334192 | 6.391415 | 0.045772 | 35.308085 | 138.360657 | 0.994027 | 3.188267 | 0.489847 | 10.514267 | 5.877909 |
| std | 0.843868 | 0.100795 | 0.121020 | 5.072058 | 0.021848 | 17.007137 | 42.498065 | 0.002991 | 0.151001 | 0.114126 | 1.230621 | 0.885639 |
| min | 3.800000 | 0.080000 | 0.000000 | 0.600000 | 0.009000 | 2.000000 | 9.000000 | 0.987110 | 2.720000 | 0.220000 | 8.000000 | 3.000000 |
| 25% | 6.300000 | 0.210000 | 0.270000 | 1.700000 | 0.036000 | 23.000000 | 108.000000 | 0.991723 | 3.090000 | 0.410000 | 9.500000 | 5.000000 |
| 50% | 6.800000 | 0.260000 | 0.320000 | 5.200000 | 0.043000 | 34.000000 | 134.000000 | 0.993740 | 3.180000 | 0.470000 | 10.400000 | 6.000000 |
| 75% | 7.300000 | 0.320000 | 0.390000 | 9.900000 | 0.050000 | 46.000000 | 167.000000 | 0.996100 | 3.280000 | 0.550000 | 11.400000 | 6.000000 |
| max | 14.200000 | 1.100000 | 1.660000 | 65.800000 | 0.346000 | 289.000000 | 440.000000 | 1.038980 | 3.820000 | 1.080000 | 14.200000 | 9.000000 |
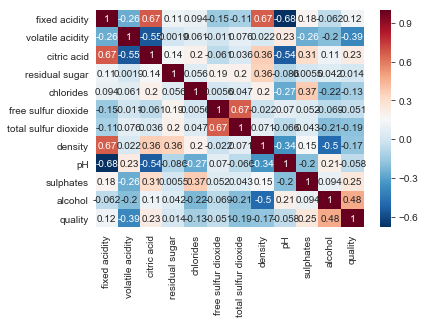
correlations_red = ex_red.corr()
correlations_white = ex_white.corr()
plt.figsize = (10,9)
sns.heatmap(correlations_red, annot=True, cmap='RdBu_r')
<matplotlib.axes._subplots.AxesSubplot at 0x1a1c7dc748>
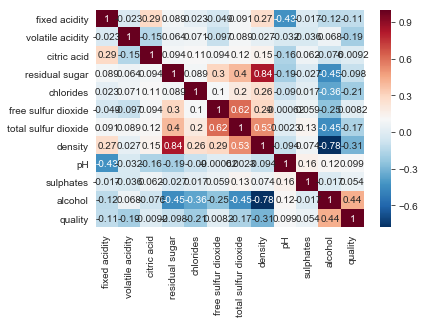
plt.figsize = (10,9)
sns.heatmap(correlations_white, annot=True, cmap='RdBu_r')
<matplotlib.axes._subplots.AxesSubplot at 0x1a1d1c9828>
sns.violinplot(ex_red.quality)
/anaconda3/lib/python3.7/site-packages/scipy/stats/stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval
<matplotlib.axes._subplots.AxesSubplot at 0x1a1cfe2e80>
sns.violinplot(ex_white.quality)
<matplotlib.axes._subplots.AxesSubplot at 0x1a1d56e828>
II. Generate Analytical Base Table
red = pd.read_csv('winequality-red.csv', sep=';')
white = pd.read_csv('winequality-white.csv', sep=';')
III. Tune Models and Select Winning Algorithm
We will train the red models first and then the white.
Regression on Red Wines
y = red.quality
X = red.drop('quality', axis=1)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = .2, random_state=1234)
print(X_train.shape, X_test.shape, y_train.shape, y_test.shape)
(1279, 11) (320, 11) (1279,) (320,)
pipelines = {
'lasso' : make_pipeline(StandardScaler(), Lasso(random_state=123)),
'ridge' : make_pipeline(StandardScaler(), Ridge(random_state=123)),
'enet' : make_pipeline(StandardScaler(), ElasticNet(random_state=123)),
'rf' : make_pipeline(StandardScaler(), RandomForestRegressor(random_state=123)),
'gb' : make_pipeline(StandardScaler(), GradientBoostingRegressor(random_state=123))
}
lasso_hyperparameters = {
'lasso__alpha' : [0.0001, 0.001, 0.01, 0.1, 1, 5, 10]
}
ridge_hyperparameters = {
'ridge__alpha' : [0.0001, 0.001, 0.01, 0.1, 1, 5, 10]
}
enet_hyperparameters = {
'elasticnet__alpha' : [0.0001, 0.001, 0.01, 0.1, 1, 5, 10],
'elasticnet__l1_ratio' : [0.1, 0.3, 0.5, 0.7, 0.9]
}
rf_hyperparameters = {
'randomforestregressor__n_estimators' : [100, 200],
'randomforestregressor__max_features' : ['auto', 'sqrt', 0.5, 0.33, 0.2]
}
gb_hyperparameters = {
'gradientboostingregressor__n_estimators' : [100, 200],
'gradientboostingregressor__learning_rate' : [0.02, 0.05, 0.1, 0.2, 0.5],
'gradientboostingregressor__max_depth': [1, 2, 3]
}
hyperparameters = {
'rf' : rf_hyperparameters,
'gb' : gb_hyperparameters,
'lasso' : lasso_hyperparameters,
'ridge' : ridge_hyperparameters,
'enet' : enet_hyperparameters
}
fitted_models = {}
for name, pipeline in pipelines.items():
model = GridSearchCV(pipeline, hyperparameters[name], cv=10, n_jobs=-1)
model.fit(X_train, y_train)
fitted_models[name] = model
print(f'{name} has been fitted.')
lasso has been fitted.
ridge has been fitted.
enet has been fitted.
rf has been fitted.
gb has been fitted.
for name, model in fitted_models.items():
print(name, type(model))
lasso <class 'sklearn.model_selection._search.GridSearchCV'>
ridge <class 'sklearn.model_selection._search.GridSearchCV'>
enet <class 'sklearn.model_selection._search.GridSearchCV'>
rf <class 'sklearn.model_selection._search.GridSearchCV'>
gb <class 'sklearn.model_selection._search.GridSearchCV'>
for name, model in fitted_models.items():
try:
model.predict(X_test)
print(f'{name} can be predicted')
except NotFittedError as e:
print(repr(e))
lasso can be predicted
ridge can be predicted
enet can be predicted
rf can be predicted
gb can be predicted
for name, model in fitted_models.items():
print(name, model.best_score_)
lasso 0.3380728789101738
ridge 0.3344467364553082
enet 0.3379619676582303
rf 0.47598436259304555
gb 0.3938200044757678
for name, model in fitted_models.items():
pred = model.predict(X_test)
print(name)
print('--------')
pred_rnd = [round(n) for n in pred]
print(f'Accuracy: {accuracy_score(y_test, pred_rnd)*100:.1f}%')
print(f'R^2: {r2_score(y_test,pred)}')
print(f'MAE: {mean_absolute_error(y_test, pred)}')
print()
lasso
--------
Accuracy: 63.4%
R^2: 0.3639837143675275
MAE: 0.47595885488079076
ridge
--------
Accuracy: 65.0%
R^2: 0.3674883892991997
MAE: 0.47054856852206195
enet
--------
Accuracy: 63.7%
R^2: 0.36429135863048057
MAE: 0.475364000358384
rf
--------
Accuracy: 75.3%
R^2: 0.5070378956609388
MAE: 0.38737499999999997
gb
--------
Accuracy: 66.9%
R^2: 0.3920069033424246
MAE: 0.4488029793340935
Regression on White Wines
y = white.quality
X = white.drop('quality', axis=1)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = .2, random_state=1234)
print(X_train.shape, X_test.shape, y_train.shape, y_test.shape)
(3918, 11) (980, 11) (3918,) (980,)
pipelines = {
'lasso' : make_pipeline(StandardScaler(), Lasso(random_state=123)),
'ridge' : make_pipeline(StandardScaler(), Ridge(random_state=123)),
'enet' : make_pipeline(StandardScaler(), ElasticNet(random_state=123)),
'rf' : make_pipeline(StandardScaler(), RandomForestRegressor(random_state=123)),
'gb' : make_pipeline(StandardScaler(), GradientBoostingRegressor(random_state=123))
}
lasso_hyperparameters = {
'lasso__alpha' : [0.0001, 0.001, 0.01, 0.1, 1, 5, 10]
}
ridge_hyperparameters = {
'ridge__alpha' : [0.0001, 0.001, 0.01, 0.1, 1, 5, 10]
}
enet_hyperparameters = {
'elasticnet__alpha' : [0.0001, 0.001, 0.01, 0.1, 1, 5, 10],
'elasticnet__l1_ratio' : [0.1, 0.3, 0.5, 0.7, 0.9]
}
rf_hyperparameters = {
'randomforestregressor__n_estimators' : [100, 200],
'randomforestregressor__max_features' : ['auto', 'sqrt', 0.5, 0.33, 0.2]
}
gb_hyperparameters = {
'gradientboostingregressor__n_estimators' : [100, 200],
'gradientboostingregressor__learning_rate' : [0.02, 0.05, 0.1, 0.2, 0.5],
'gradientboostingregressor__max_depth': [1, 2, 3]
}
hyperparameters = {
'rf' : rf_hyperparameters,
'gb' : gb_hyperparameters,
'lasso' : lasso_hyperparameters,
'ridge' : ridge_hyperparameters,
'enet' : enet_hyperparameters
}
fitted_models = {}
for name, pipeline in pipelines.items():
model = GridSearchCV(pipeline, hyperparameters[name], cv=10, n_jobs=-1)
model.fit(X_train, y_train)
fitted_models[name] = model
print(f'{name} has been fitted.')
lasso has been fitted.
ridge has been fitted.
enet has been fitted.
rf has been fitted.
gb has been fitted.
for name, model in fitted_models.items():
print(name, type(model))
lasso <class 'sklearn.model_selection._search.GridSearchCV'>
ridge <class 'sklearn.model_selection._search.GridSearchCV'>
enet <class 'sklearn.model_selection._search.GridSearchCV'>
rf <class 'sklearn.model_selection._search.GridSearchCV'>
gb <class 'sklearn.model_selection._search.GridSearchCV'>
for name, model in fitted_models.items():
try:
model.predict(X_test)
print(f'{name} can be predicted')
except NotFittedError as e:
print(repr(e))
lasso can be predicted
ridge can be predicted
enet can be predicted
rf can be predicted
gb can be predicted
for name, model in fitted_models.items():
print(name, model.best_score_)
lasso 0.2845506011036594
ridge 0.2845935195724066
enet 0.28458806763285216
rf 0.5303098868482835
gb 0.4208457648616886
for name, model in fitted_models.items():
pred = model.predict(X_test)
print(name)
print('--------')
pred_rnd = [round(n) for n in pred]
print(f'Accuracy: {accuracy_score(y_test, pred_rnd)*100:.1f}%')
print(f'R^2: {r2_score(y_test,pred)}')
print(f'MAE: {mean_absolute_error(y_test, pred)}')
print()
lasso
--------
Accuracy: 53.1%
R^2: 0.22591469935875141
MAE: 0.5898414215829592
ridge
--------
Accuracy: 51.9%
R^2: 0.2247048691371496
MAE: 0.5898967574235963
enet
--------
Accuracy: 52.0%
R^2: 0.22396523747696395
MAE: 0.5899893127993073
rf
--------
Accuracy: 69.9%
R^2: 0.5133021260245902
MAE: 0.42600000000000005
gb
--------
Accuracy: 62.4%
R^2: 0.38960332183177027
MAE: 0.5159582270274666
Binary Classification on Reds
def sort_good(quality):
good = quality>=7
return good
y = red.quality.apply(sort_good)
X = red.drop('quality', axis=1)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = .2, random_state=1234)
print(X_train.shape, X_test.shape, y_train.shape, y_test.shape)
(1279, 11) (320, 11) (1279,) (320,)
print(y[5:10])
5 False
6 False
7 True
8 True
9 False
Name: quality, dtype: bool
pipelines = {
'rf' : make_pipeline(StandardScaler(), RandomForestClassifier(random_state=123)),
'gb' : make_pipeline(StandardScaler(), GradientBoostingClassifier(random_state=123))
}
rf_hyperparameters = {
'randomforestclassifier__n_estimators' : [50, 100, 200],
'randomforestclassifier__max_features' : ['auto', 0.5, 0.33, 0.2]
}
gb_hyperparameters = {
'gradientboostingclassifier__n_estimators' : [50, 100, 200],
'gradientboostingclassifier__learning_rate' : [0.02, 0.05, 0.1, 0.2, 0.5],
'gradientboostingclassifier__max_depth': [1, 2, 3, 5]
}
hyperparameters = {
'rf' : rf_hyperparameters,
'gb' : gb_hyperparameters,
}
fitted_models = {}
for name, pipeline in pipelines.items():
model = GridSearchCV(pipeline, hyperparameters[name], cv=10, n_jobs=-1)
model.fit(X_train, y_train)
fitted_models[name] = model
print(f'{name} has been fitted.')
rf has been fitted.
gb has been fitted.
for name, model in fitted_models.items():
print(name, type(model))
rf <class 'sklearn.model_selection._search.GridSearchCV'>
gb <class 'sklearn.model_selection._search.GridSearchCV'>
for name, model in fitted_models.items():
try:
model.predict(X_test)
print(f'{name} can be predicted')
except NotFittedError as e:
print(repr(e))
rf can be predicted
gb can be predicted
for name, model in fitted_models.items():
print(name, model.best_score_)
rf 0.910086004691165
gb 0.9093041438623924
for name, model in fitted_models.items():
pred = model.predict(X_test)
print(name)
print('--------')
pred_prob = model.predict_proba(X_test)
pred_prob = [p[1] for p in pred_prob]
print(f'Accuracy: {accuracy_score(y_test, pred)*100:.1f}%')
print(f'AUROC: {roc_auc_score(y_test, pred_prob)}')
print()
rf
--------
Accuracy: 90.0%
AUROC: 0.9130168721042049
gb
--------
Accuracy: 90.0%
AUROC: 0.9143281755398199
Binary Classification on Whites
y = white.quality.apply(sort_good)
X = white.drop('quality', axis=1)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = .2, random_state=1234)
print(X_train.shape, X_test.shape, y_train.shape, y_test.shape)
(3918, 11) (980, 11) (3918,) (980,)
print(y[5:10])
5 False
6 False
7 False
8 False
9 False
Name: quality, dtype: bool
pipelines = {
'rf' : make_pipeline(StandardScaler(), RandomForestClassifier(random_state=123)),
'gb' : make_pipeline(StandardScaler(), GradientBoostingClassifier(random_state=123))
}
rf_hyperparameters = {
'randomforestclassifier__n_estimators' : [50, 100, 200],
'randomforestclassifier__max_features' : ['auto', 0.5, 0.33, 0.2]
}
gb_hyperparameters = {
'gradientboostingclassifier__n_estimators' : [50, 100, 200],
'gradientboostingclassifier__learning_rate' : [0.02, 0.05, 0.1, 0.2, 0.5],
'gradientboostingclassifier__max_depth': [1, 2, 3, 5]
}
hyperparameters = {
'rf' : rf_hyperparameters,
'gb' : gb_hyperparameters,
}
fitted_models = {}
for name, pipeline in pipelines.items():
model = GridSearchCV(pipeline, hyperparameters[name], cv=10, n_jobs=-1)
model.fit(X_train, y_train)
fitted_models[name] = model
print(f'{name} has been fitted.')
rf has been fitted.
gb has been fitted.
for name, model in fitted_models.items():
print(name, type(model))
rf <class 'sklearn.model_selection._search.GridSearchCV'>
gb <class 'sklearn.model_selection._search.GridSearchCV'>
for name, model in fitted_models.items():
try:
model.predict(X_test)
print(f'{name} can be predicted')
except NotFittedError as e:
print(repr(e))
rf can be predicted
gb can be predicted
for name, model in fitted_models.items():
print(name, model.best_score_)
rf 0.8754466564573762
gb 0.8670239918325676
for name, model in fitted_models.items():
pred = model.predict(X_test)
print(name)
print('--------')
pred_prob = model.predict_proba(X_test)
pred_prob = [p[1] for p in pred_prob]
print(f'Accuracy: {accuracy_score(y_test, pred)*100:.1f}%')
print(f'AUROC: {roc_auc_score(y_test, pred_prob)}')
print()
rf
--------
Accuracy: 88.6%
AUROC: 0.9093720095693779
gb
--------
Accuracy: 85.8%
AUROC: 0.8918779904306219
IV. Analysis
I was not able to achieve my first win condition of predicting the integer value of the win qualities to 90% accuracy, I was able to acheive the classification win condition of predicting good wines with an AUROC score above .9.
# Model for best Red Wines Model
fitted_models['gb'].best_estimator_.named_steps['gradientboostingclassifier']
GradientBoostingClassifier(criterion='friedman_mse', init=None,
learning_rate=0.05, loss='deviance', max_depth=5,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=200,
n_iter_no_change=None, presort='auto', random_state=123,
subsample=1.0, tol=0.0001, validation_fraction=0.1,
verbose=0, warm_start=False)
# Model for best White Wines Model
fitted_models['rf'].best_estimator_.named_steps['randomforestclassifier']
RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=200, n_jobs=None,
oob_score=False, random_state=123, verbose=0, warm_start=False)